Showing 1 to 15 of 2075 results


New fire resistant plaster based material
Patents for licensing UNIVERSIDAD DE BURGOS

New device for bidirectional optical link
Patents for licensing Universitat Politècnica de Catalunya - UPC

New quinoline compounds for use in the treatment and prevention of viral
Patents for licensing CSIC - Consejo Superior de Investigaciones Científicas

Gelled microspheres for the vehiculation and release of therapeutic agents
Patents for licensing Universidad de Granada

Magnetic resonance imaging (MRI) Of Tissue Fibrosis Without Contrast Agent
Patents for licensing Yeda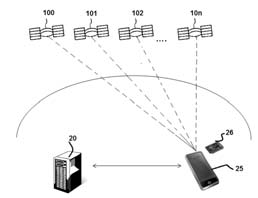

Method and system for correcting errors in satellite positioning systems and computer program products thereof
Patents for licensing Universitat Politècnica de Catalunya - UPC![A new method of obtaining mixoxanthophylls from Arthrospira platensis dye extracts (from spirulina) for use in the pharmaceutical […]](https://static6.innoget.com/uploads/88e3c8b21489c0f2a98efd73be95b10d4e5f691f.jpg)

A new method of obtaining mixoxanthophylls from Arthrospira platensis dye extracts (from spirulina) for use in the pharmaceutical […]
Patents for licensing Jagiellonian University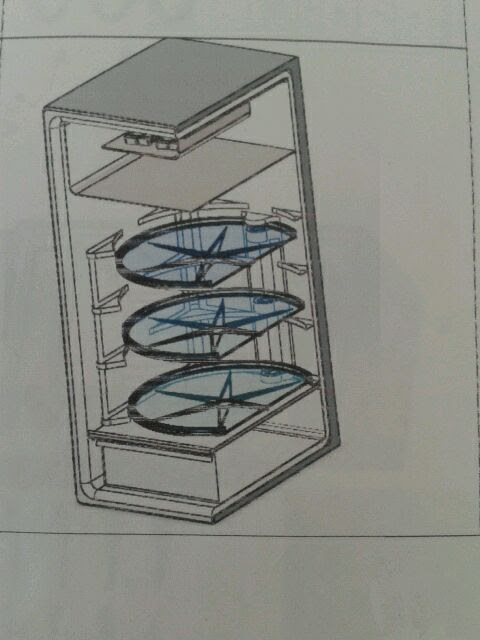

Mechanism drive system for activating rotating trays of household appliances and the like.
Innovative Products and Technologies BATHINNOVAX

Dispensing system for flexible packaging
Innovative Products and Technologies impulse

Low cost sensors for the detection of gaseous hydrogen
Patents for licensing UNIVERSIDAD DE ALICANTE

Demetalization of sewage sludges through physico-chemical processes
Research Services and Capabilities UNIVERSIDAD DE BURGOS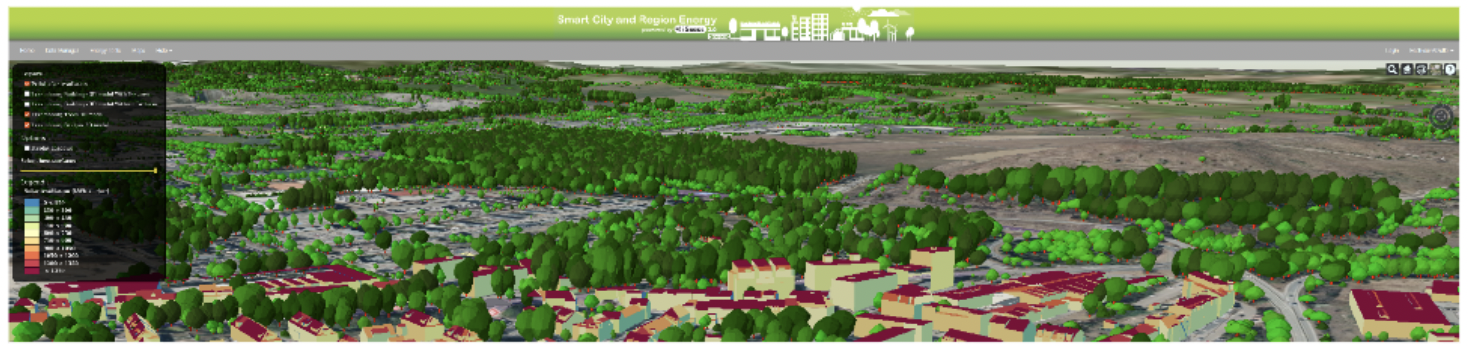

iGuess4SECAP - An Energy Transition Platform for Sustainable Energy and Climate Action Planning for Cities and Regions
Innovative Products and Technologies Luxembourg Institute of Science and Technology (LIST)

Scalable burst assembly algorithm for Optical Burst Switching networks
Patents for licensing Universitat Politècnica de Catalunya - UPC

SERS-active substrates based on plasmon nanomaterials
Innovative Products and Technologies BSUIR R&D Department

